Quickstart#
hictkpy provides Python bindings for hictk through pybind11.
hictk.File() can open .cool and .hic files and allows retrieval of interactions as well as file metadata.
The example use file 4DNFIOTPSS3L.hic, which can be downloaded from here.
Opening files#
In [1]: import hictkpy as htk
# .mcool and .cool files work as well
In [2]: f = htk.File("4DNFIOTPSS3L.hic", 10_000)
In [3]: f.path()
Out[3]: '4DNFIOTPSS3L.hic'
Reading file metadata#
In [4]: f.bin_size()
Out[4]: 10000
In [5]: f.chromosomes()
Out[5]:
{'2L': 23513712,
'2R': 25286936,
'3L': 28110227,
'3R': 32079331,
'4': 1348131,
'X': 23542271,
'Y': 3667352}
In [6]: f.attributes()
Out[6]:
{'bin_size': 10000,
'format': 'HIC',
'format_version': 8,
'assembly': '/var/lib/cwl/stgb25a903a-ebb6-4a56-bf3f-90bd84a40bf4/4DNFIBEEN92C.chrom.sizes',
'format-url': 'https://github.com/aidenlab/hic-format',
'nbins': 13758,
'nchroms': 8}
Fetch interactions#
Interactions can be fetched by calling the hictkpy.File.fetch() method on hictkpy.File() objects.
hictkpy.File.fetch() returns hictkpy.PixelSelector() objects, which are very cheap to create.
# Fetch all interactions (genome-wide query) in COO format (row, column, count)
In [7]: sel = f.fetch()
# Fetch all interactions (genome-wide query) in bedgraph2 format
In [8]: sel = f.fetch(join=True)
# Fetch KR-normalized interactions
In [9]: sel = f.fetch(normalization="KR")
# Fetch interactions for a region of interest
In [9]: sel = f.fetch("2L:10,000,000-20,000,000")
In [10]: sel = f.fetch("2L:10,000,000-20,000,000", "X")
In [11]: sel.nnz()
Out[11]: 2247057
In [12]: sel.sum()
Out[12]: 7163361
Fetching interactions as pandas DataFrames#
In [13]: sel = f.fetch("2L:10,000,000-20,000,000", join=True)
In [14]: sel.to_df()
Out[14]:
chrom1 start1 end1 chrom2 start2 end2 count
0 2L 10000000 10010000 2L 10000000 10010000 6759
1 2L 10000000 10010000 2L 10010000 10020000 3241
2 2L 10000000 10010000 2L 10020000 10030000 760
3 2L 10000000 10010000 2L 10030000 10040000 454
4 2L 10000000 10010000 2L 10040000 10050000 289
... ... ... ... ... ... ... ...
339036 2L 19970000 19980000 2L 19980000 19990000 407
339037 2L 19970000 19980000 2L 19990000 20000000 221
339038 2L 19980000 19990000 2L 19980000 19990000 391
339039 2L 19980000 19990000 2L 19990000 20000000 252
339040 2L 19990000 20000000 2L 19990000 20000000 266
[339041 rows x 7 columns]
Fetching interactions as scipy.sparse.coo_matrix#
In [15]: sel = f.fetch("2L:10,000,000-20,000,000", join=True)
In [16]: sel.to_coo()
Out[16]:
<1000x1000 sparse matrix of type '<class 'numpy.int32'>'
with 339041 stored elements in COOrdinate format>
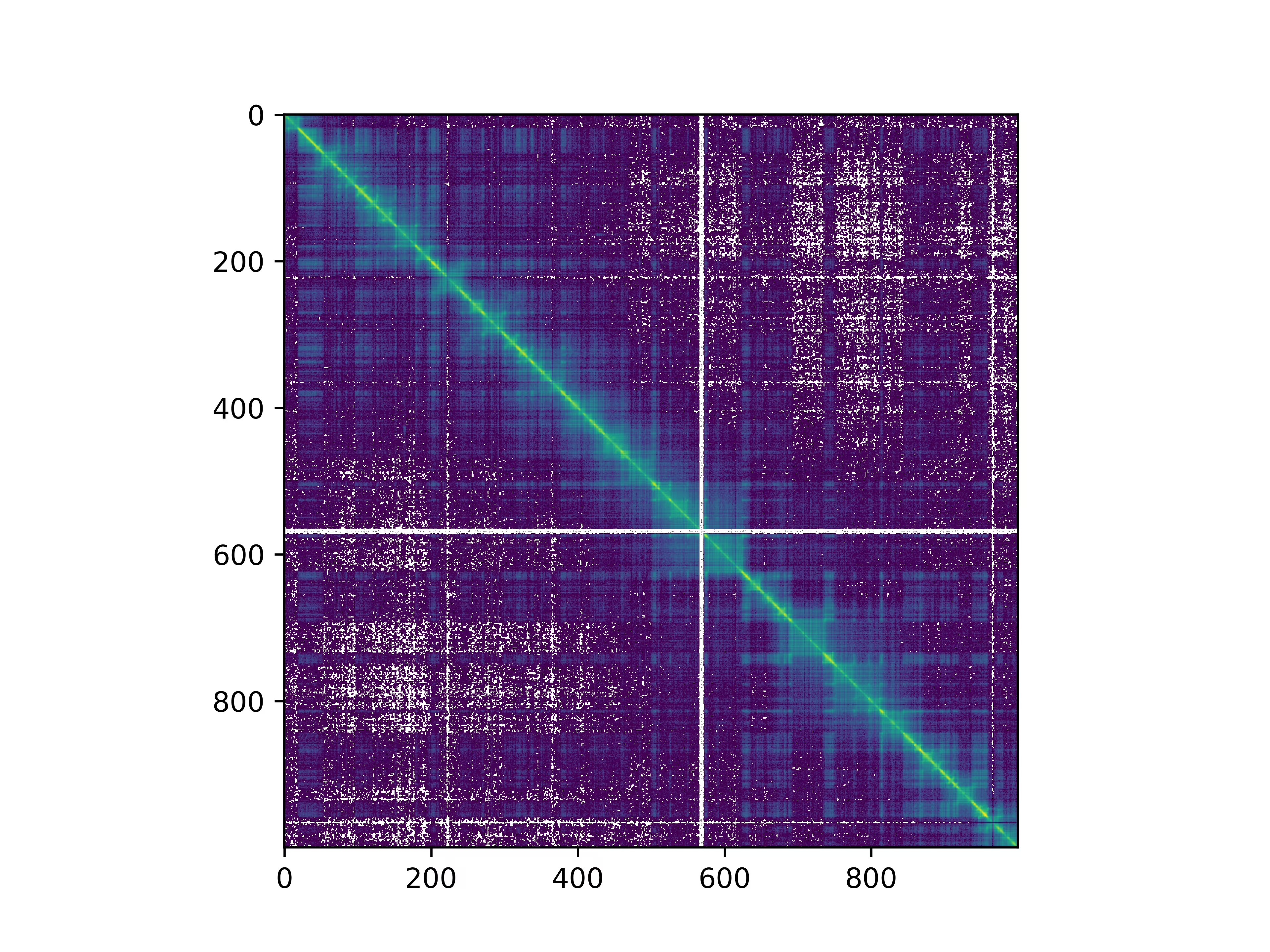
Fetching interactions as numpy NDarray#
In [17]: sel = f.fetch("2L:10,000,000-20,000,000", join=True)
In [18]: m = sel.to_numpy()
In [19]: import matplotlib.pyplot as plt
In [20]: from matplotlib.colors import LogNorm
In [21]: plt.imshow(m, norm=LogNorm())
In [22]: plt.show()